

Next:
6 Discussion and future
Up:
Robust Contour Tracking in
Previous:
4 Improving feature detection
The ultimate goal of this work is to demonstrate that automated image
analysis can be used for to perform temporal-based quantification of
regional heart function. As a step towards this goal, in this section we
present some results of applying the training and tracking procedures
outlined in Section
3
to some further real heart image sequences.
Abnormal heart motion was simulated by editing images corresponding to
the diastole section of the cardiac cycle for a normal heart image
sequence (the example used in Section
3
) so that the posterior (left) wall appeared sluggish. This was done by
shifting an image block containing the posterior wall from each of the
diastole images by 10 pixels to the right. The image block was then
blended in with the data using an exponential weight function. Finally a
PCA was performed as before. Figure
6
summarises the results. The key thing to observe is that the nature of
the principal modes remain unchanged although the magnitude is affected
(compare with Figure
1
). In particular the second mode (middle plot) shows that the posterior
wall scales outwards to a lesser degree which is consistent with the
imposed abnormality.

Figure 6:
Principal component analysis performed on 4 cardiac cycles of a
ultrasonic image sequence. The actual data (light curve) is plotted
along with the simulated data (dark curve). The mean shape (left) is
plotted. The modes represent the addition of standard deviations to the mean shape; From second left mode 1 (the
dominant mode) to mode 4.
standard deviations to the mean shape; From second left mode 1 (the
dominant mode) to mode 4.
A PCA was performed on four manually segmented non-consecutive cycles of
real data for a patient diagnosed with a disease which manifests as a
loss in elasticity of the heart. Figure
7
summarises the results of the PCA. In this case seven modes of variation
express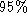 of the variability as compared to four modes with the normal heart. The
first mode appears to be a scaling of the anterior wall, the second mode
a translation mode, the third mode a scaling and the fourth mode a
mixture of a translation and scaling. The fifth mode is translation.
of the variability as compared to four modes with the normal heart. The
first mode appears to be a scaling of the anterior wall, the second mode
a translation mode, the third mode a scaling and the fourth mode a
mixture of a translation and scaling. The fifth mode is translation.
|
Mode
|
Eigenvalue
|
Variability percent
|
Cumulative variability
|
|
1
|
669.51
|
0.321
|
0.321
|
|
2
|
544.00
|
0.294
|
0.615
|
|
3
|
284.00
|
0.139
|
0.755
|
|
4
|
169.4
|
0.115
|
0.871
|
|
5
|
85.29
|
0.0546
|
0.925
|
|
6
|
45.54
|
0.0185
|
0.944
|
|
7
|
31.35
|
0.0139
|
0.958
|
Table 3:
The results of applying a principal component analysis to 4 manually
segmented cardiac cycles from an abnormal heart. It is clear that 7
modes of variation explain over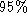 of the variability.
of the variability.

Figure 7:
First five principal modes for an abnormal heart. The mean shape (filled
line) is plotted along with flow lines representing how the start of
each span behaves with the addition of standard deviations to the mean. From left, mode 1 (the dominant mode)
to mode 5.
standard deviations to the mean. From left, mode 1 (the dominant mode)
to mode 5.


Next:
6 Discussion and future
Up:
Robust Contour Tracking in
Previous:
4 Improving feature detection
Gary Jacob
Tue Jul 22 17:45:09 BST 1997

 standard deviations to the mean shape; From second left mode 1 (the
dominant mode) to mode 4.
standard deviations to the mean shape; From second left mode 1 (the
dominant mode) to mode 4.


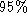 of the variability as compared to four modes with the normal heart. The
first mode appears to be a scaling of the anterior wall, the second mode
a translation mode, the third mode a scaling and the fourth mode a
mixture of a translation and scaling. The fifth mode is translation.
of the variability as compared to four modes with the normal heart. The
first mode appears to be a scaling of the anterior wall, the second mode
a translation mode, the third mode a scaling and the fourth mode a
mixture of a translation and scaling. The fifth mode is translation.